Note
Click here to download the full example code
Reconstruct with Diffusion Spectrum Imaging
We show how to apply Diffusion Spectrum Imaging [Wedeen08] to diffusion MRI datasets of Cartesian keyhole diffusion gradients.
First import the necessary modules:
import numpy as np
from dipy.core.gradients import gradient_table
from dipy.data import get_fnames, get_sphere
from dipy.io.gradients import read_bvals_bvecs
from dipy.io.image import load_nifti
from dipy.reconst.dsi import DiffusionSpectrumModel
Download and get the data filenames for this tutorial.
fraw, fbval, fbvec = get_fnames('taiwan_ntu_dsi')
img contains a nibabel Nifti1Image object (data) and gtab contains a GradientTable object (gradient information e.g. b-values). For example to read the b-values it is possible to write print(gtab.bvals).
Load the raw diffusion data and the affine.
data, affine, voxel_size = load_nifti(fraw, return_voxsize=True)
bvals, bvecs = read_bvals_bvecs(fbval, fbvec)
bvecs[1:] = (bvecs[1:] /
np.sqrt(np.sum(bvecs[1:] * bvecs[1:], axis=1))[:, None])
gtab = gradient_table(bvals, bvecs)
print('data.shape (%d, %d, %d, %d)' % data.shape)
data.shape (96, 96, 60, 203)
data.shape (96, 96, 60, 203)
This dataset has anisotropic voxel sizes, therefore reslicing is necessary.
Instantiate the Model and apply it to the data.
dsmodel = DiffusionSpectrumModel(gtab)
Let’s just use one slice only from the data.
dataslice = data[:, :, data.shape[2] // 2]
dsfit = dsmodel.fit(dataslice)
0%| | 0/9216 [00:00<?, ?it/s]
100%|##########| 9216/9216 [00:00<00:00, 595622.45it/s]
Load an odf reconstruction sphere
sphere = get_sphere('repulsion724')
Calculate the ODFs with this specific sphere
ODF = dsfit.odf(sphere)
print('ODF.shape (%d, %d, %d)' % ODF.shape)
/Users/skoudoro/devel/dipy/dipy/reconst/dsi.py:173: RuntimeWarning: invalid value encountered in divide
Pr /= Pr.sum()
ODF.shape (96, 96, 724)
ODF.shape (96, 96, 724)
In a similar fashion it is possible to calculate the PDFs of all voxels in one call with the following way
PDF = dsfit.pdf()
print('PDF.shape (%d, %d, %d, %d, %d)' % PDF.shape)
PDF.shape (96, 96, 17, 17, 17)
PDF.shape (96, 96, 17, 17, 17)
We see that even for a single slice this PDF array is close to 345 MBytes so we really have to be careful with memory usage when use this function with a full dataset.
The simple solution is to generate/analyze the ODFs/PDFs by iterating through each voxel and not store them in memory if that is not necessary.
from dipy.core.ndindex import ndindex
for index in ndindex(dataslice.shape[:2]):
pdf = dsmodel.fit(dataslice[index]).pdf()
If you really want to save the PDFs of a full dataset on the disc we recommend
using memory maps (numpy.memmap) but still have in mind that even if you do
that for example for a dataset of volume size (96, 96, 60) you will need
about 2.5 GBytes which can take less space when reasonable spheres
(with < 1000 vertices) are used.
Let’s now calculate a map of Generalized Fractional Anisotropy (GFA) [Tuch04] using the DSI ODFs.
from dipy.reconst.odf import gfa
GFA = gfa(ODF)
import matplotlib.pyplot as plt
fig_hist, ax = plt.subplots(1)
ax.set_axis_off()
plt.imshow(GFA.T)
plt.savefig('dsi_gfa.png', bbox_inches='tight')
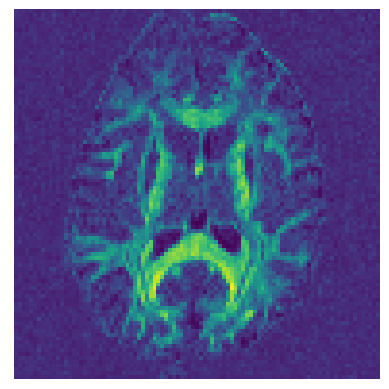

See also example_reconst_dsi_metrics for calculating different types of DSI maps.
Wedeen et al., Diffusion spectrum magnetic resonance imaging (DSI) tractography of crossing fibers, Neuroimage, vol 41, no 4, 1267-1277, 2008.
Tuch, D.S, Q-ball imaging, MRM, vol 52, no 6, 1358-1372, 2004.
Total running time of the script: ( 0 minutes 24.601 seconds)