Note
Click here to download the full example code
Surface seeding for tractography
Surface seeding is a way to generate initial position for tractography from cortical surfaces position [Stonge2018].
import numpy as np
from dipy.viz import window, actor
from dipy.data import get_fnames
from dipy.tracking.mesh import (random_coordinates_from_surface,
seeds_from_surface_coordinates)
from fury.io import load_polydata
from fury.utils import (get_polydata_triangles, get_polydata_vertices,
get_actor_from_polydata, normals_from_v_f)
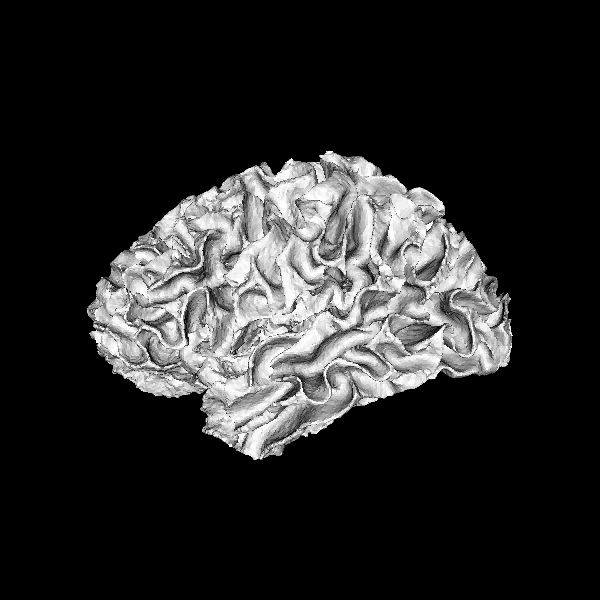
Fetch and load a surface
brain_lh = get_fnames("fury_surface")
polydata = load_polydata(brain_lh)
Extract the triangles and vertices
triangles = get_polydata_triangles(polydata)
vts = get_polydata_vertices(polydata)
Display the surface
First, create an actor from the polydata, to display in the scene
scene = window.Scene()
surface_actor = get_actor_from_polydata(polydata)
scene.add(surface_actor)
scene.set_camera(position=(-500, 0, 0),
view_up=(0.0, 0.0, 1))
# Uncomment the line below to show to display the window
# window.show(scene, size=(600, 600), reset_camera=False)
window.record(scene, out_path='surface_seed1.png', size=(600, 600))

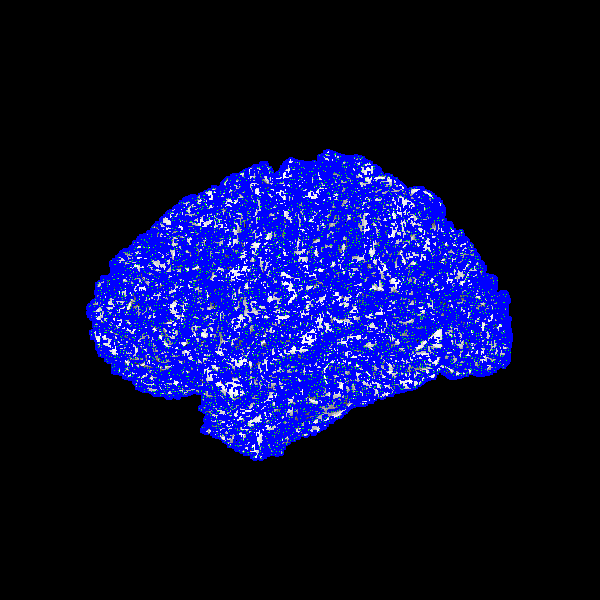
Generate a list of seeding positions
Choose the number of seed
nb_seeds = 100000
nb_triangles = len(triangles)
Get a list of triangles indices and trilinear coordinates for each seed
tri_idx, trilin_co = random_coordinates_from_surface(nb_triangles, nb_seeds)
Get the 3d cartesian position from triangles indices and trilinear coordinates
seed_pts = seeds_from_surface_coordinates(triangles, vts, tri_idx, trilin_co)
Compute normal and get the normal direction for each seeds
normals = normals_from_v_f(vts, triangles)
seed_n = seeds_from_surface_coordinates(triangles, normals, tri_idx, trilin_co)
Create dot actor for seeds (blue)
seed_actors = actor.dot(seed_pts, colors=(0, 0, 1), dot_size=4.0)
Create line actors for seeds normals (green outside, red inside)
normal_length = 0.5
normal_in = np.tile(seed_pts[:, np.newaxis, :], (1, 2, 1))
normal_out = np.tile(seed_pts[:, np.newaxis, :], (1, 2, 1))
normal_in[:, 0] -= seed_n * normal_length
normal_out[:, 1] += seed_n * normal_length
normal_in_actor = actor.line(normal_in, colors=(1, 0, 0))
normal_out_actor = actor.line(normal_out, colors=(0, 1, 0))
Visualise seeds and normals along the surface
scene = window.Scene()
scene.add(surface_actor)
scene.add(seed_actors)
scene.add(normal_in_actor)
scene.add(normal_out_actor)
scene.set_camera(position=(-500, 0, 0),
view_up=(0.0, 0.0, 1))
# Uncomment the line below to show to display the window
# window.show(scene, size=(600, 600), reset_camera=False)
window.record(scene, out_path='surface_seed2.png', size=(600, 600))

References
St-Onge, E., Daducci, A., Girard, G., & Descoteaux, M. Surface-enhanced tractography (SET). NeuroImage, 169, 524-539, 2018.
Total running time of the script: ( 0 minutes 1.188 seconds)