Note
Click here to download the full example code
Brain segmentation with median_otsu
We show how to extract brain information and mask from a b0 image using DIPY_’s
segment.mask module.
First import the necessary modules:
import numpy as np
from dipy.data import get_fnames
from dipy.io.image import load_nifti, save_nifti
from dipy.segment.mask import median_otsu
Download and read the data for this tutorial.
The scil_b0 dataset contains different data from different companies and
models. For this example, the data comes from a 1.5 Tesla Siemens MRI.
data_fnames = get_fnames('scil_b0')
data, affine = load_nifti(data_fnames[1])
data = np.squeeze(data)
Segment the brain using DIPY’s mask module.
median_otsu returns the segmented brain data and a binary mask of the
brain. It is possible to fine tune the parameters of median_otsu
(median_radius and num_pass) if extraction yields incorrect results
but the default parameters work well on most volumes. For this example,
we used 2 as median_radius and 1 as num_pass
b0_mask, mask = median_otsu(data, median_radius=2, numpass=1)
Saving the segmentation results is very easy. We need the b0_mask, and the
binary mask volumes. The affine matrix which transform the image’s coordinates
to the world coordinates is also needed. Here, we choose to save both images
in float32.
fname = 'se_1.5t'
save_nifti(fname + '_binary_mask.nii.gz', mask.astype(np.float32), affine)
save_nifti(fname + '_mask.nii.gz', b0_mask.astype(np.float32), affine)
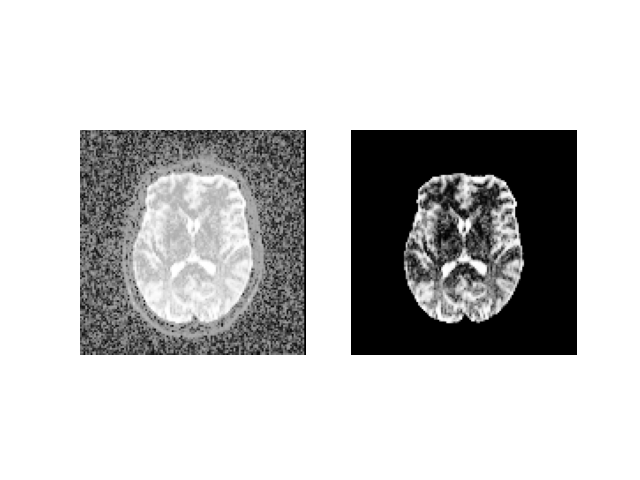
Quick view of the results middle slice using matplotlib.
import matplotlib.pyplot as plt
from dipy.core.histeq import histeq
sli = data.shape[2] // 2
plt.figure('Brain segmentation')
plt.subplot(1, 2, 1).set_axis_off()
plt.imshow(histeq(data[:, :, sli].astype('float')).T,
cmap='gray', origin='lower')
plt.subplot(1, 2, 2).set_axis_off()
plt.imshow(histeq(b0_mask[:, :, sli].astype('float')).T,
cmap='gray', origin='lower')
plt.savefig('median_otsu.png')

median_otsu can also automatically crop the outputs to remove the largest
possible number of background voxels. This makes outputted data significantly
smaller. Auto-cropping in median_otsu is activated by setting the
autocrop parameter to True.
b0_mask_crop, mask_crop = median_otsu(data, median_radius=4, numpass=4,
autocrop=True)
Saving cropped data using nibabel as demonstrated previously.
save_nifti(fname + '_binary_mask_crop.nii.gz', mask_crop.astype(np.float32),
affine)
save_nifti(fname + '_mask_crop.nii.gz', b0_mask_crop.astype(np.float32),
affine)
Total running time of the script: ( 0 minutes 21.493 seconds)